I am a computational biologist, working as a Senior Data Scientist in Lotfollahi lab in Sanger Institute. My scientific background is in bioinformatics and biochemistry, specifically interested in formulating desiderata of biology or medical science into computational problem.
“What I cannot create, I do not understand”
- Richard Feynman
Academic CV
Current research interest
•
Multi-modal learning of spatial gene expression and histopathology image
•
Uncertainty-controlled de novo regulatory sequence design
Work experience
•
Senior Data Scientist at Sanger Institute
•
Education
•
•
◦
Selected as Fellow of the National Excellence Scholarship (Natural Sciences and Engineering)
Publication
•
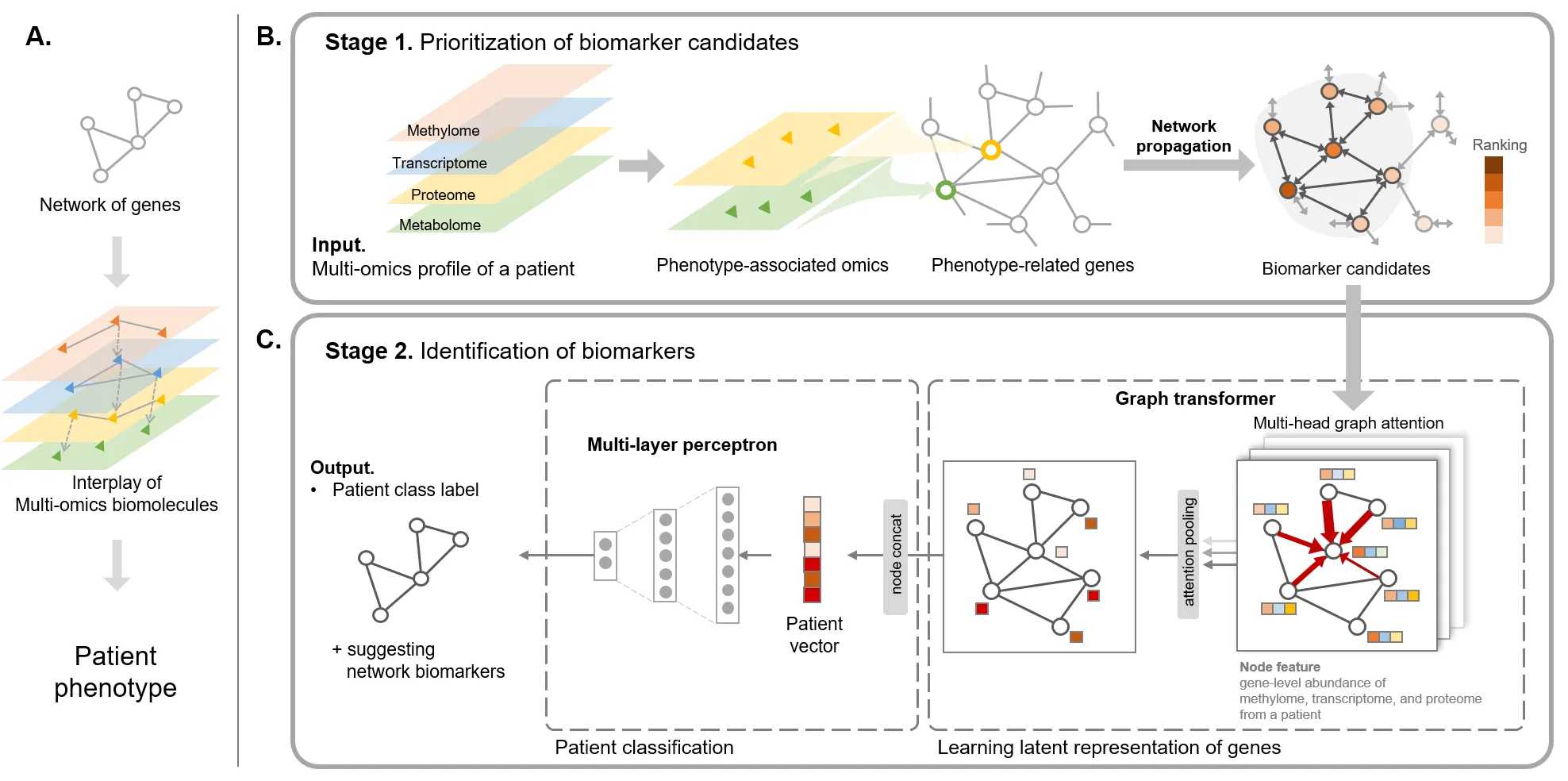
2024, BMC genomics (Under revision) “ALPACA: A Visual Data Mining System for Subcellular Location-specific Knowledge Mining from Multi-Omics Data in Cancer”
•
Co-lead 2025, BioData Mining “Identification of Severity Related Mutation Hotspots in SARS-CoV-2 Using a Density-Based Clustering Approach”
•
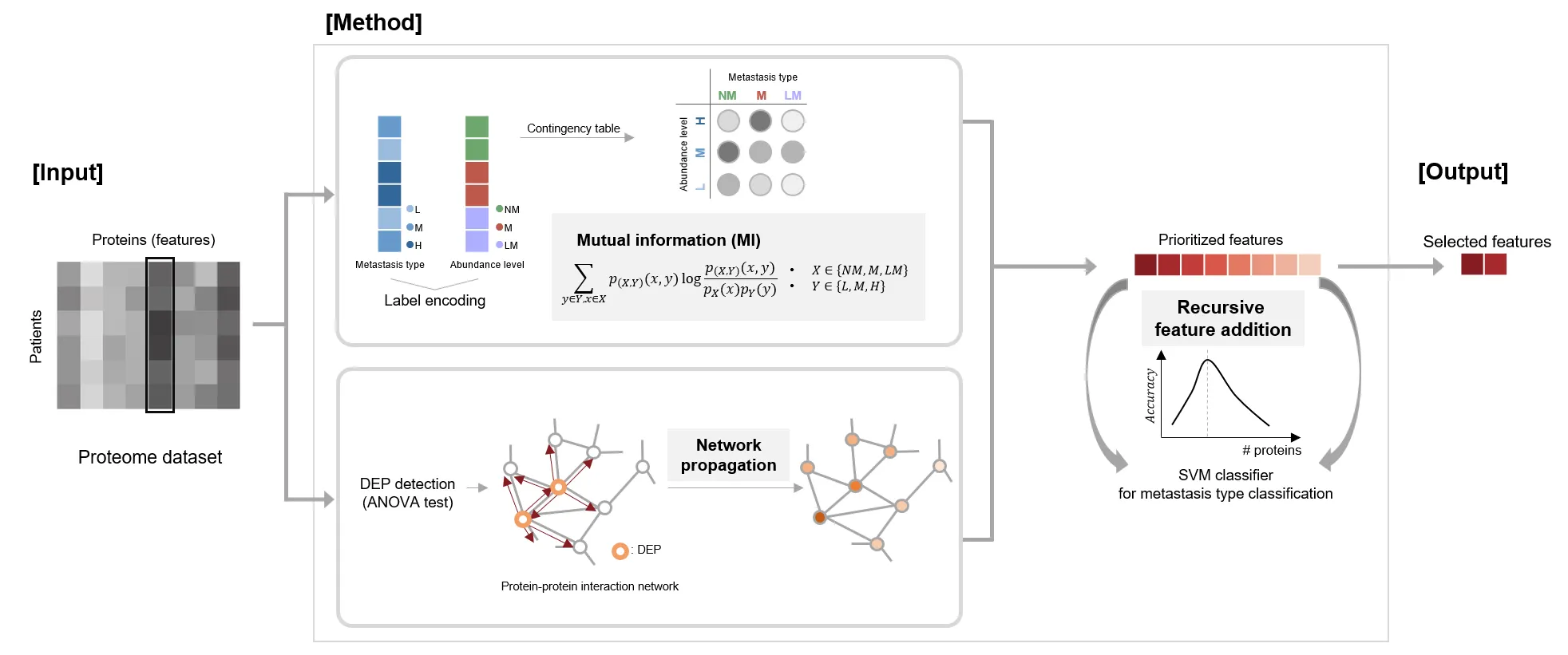
Co-lead 2024, Scientific Reports “Identification of VWA5A as a Novel Biomarker for Inhibiting Metastasis in Breast Cancer by Machine-learning Based Protein Prioritization”
•
2023, Proceedings of the 14th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics “Deep learning-based survival prediction using DNA methylation-derived 3D genomic information”
Presented by Dabin Jeong at ACM-BCB 2023
•
2023, Journal of Korean Medical Science “Machine Learning-Based Proteomics Reveals Ferroptosis in COPD Patient-Derived Airway Epithelial Cells Upon Smoking Exposure”
•
2022, International Journal of Molecular Science “A Survey on Computational Methods for Investigation on ncRNA-Disease Association through the Mode of Action Perspective”
•
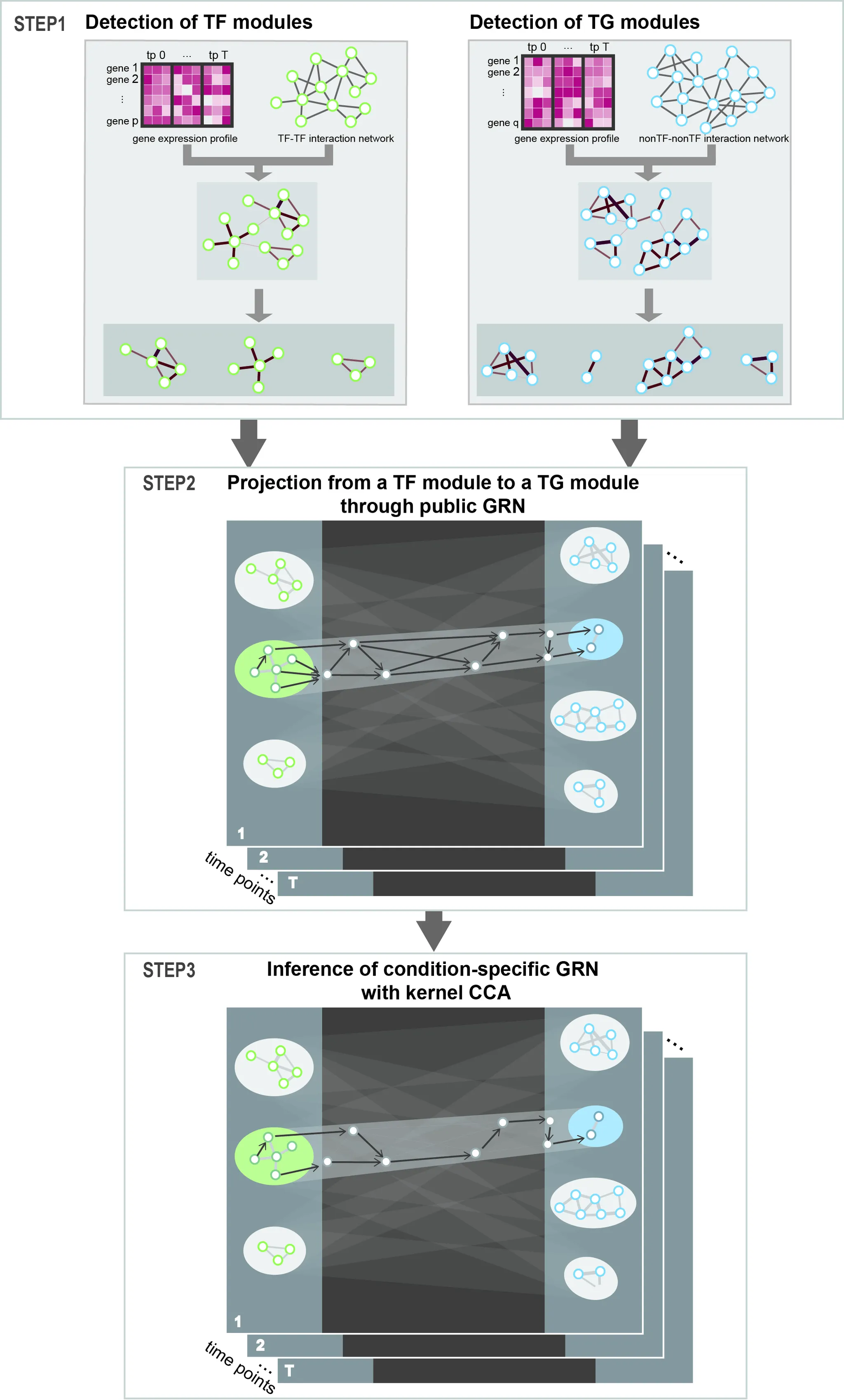
Lead 2021, Frontiers in Genetics “Construction of Condition-Specific Gene Regulatory Network using Kernel Canonical Correlation Analysis”
•
Co-lead 2020, Recent Advances in Biological network Analysis, “Network Propagation for the Analysis of Multi-omics Data”
•
Presented by Dabin Jeong at Genome Informatics Workshop (GIW) 2019
•
2019, Frontiers in Plant Science “PropaNet: Time-Varying Condition-Specific Transcriptional Network Construction by Network Propagation”
•
2018, Nucleic Acids Research “TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions”
cited 1,831 times (25.9.11)
Portfolio
Skills
•
Python
•
R
•
Docker
•
Nextflow
•
Snakemake
•
Git
•
Shell script