Goal
cell representation learning
Task (in ML perspective)
clustering
Challenges
•
Dropout events
•
Embedding for clustering
Methods
•
Dataset
◦
Multiple modalities
▪
scCITE-seq : mRNA + ADT
▪
scSMAGE-seq: mRNA + ATAC
▪
scATAC-seq
▪
scRNA-seq
•
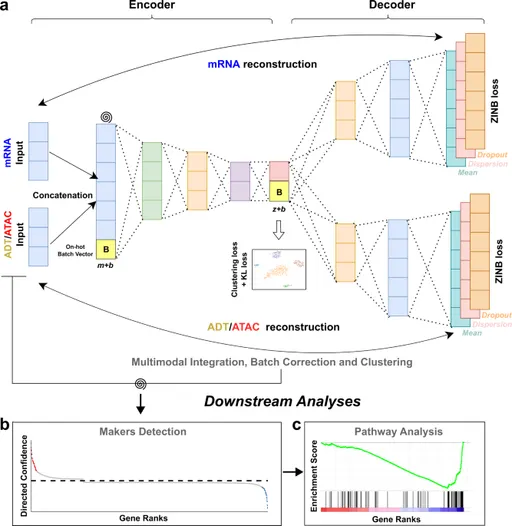
Method
◦
input : CITE-seq or SMAGE-seq dataset
◦
output :
◦
Detailed method
Reconstruction loss: ZINB (Zero enflated negative binomial) loss
KL loss
K-means Clustering loss
▪
최종 loss
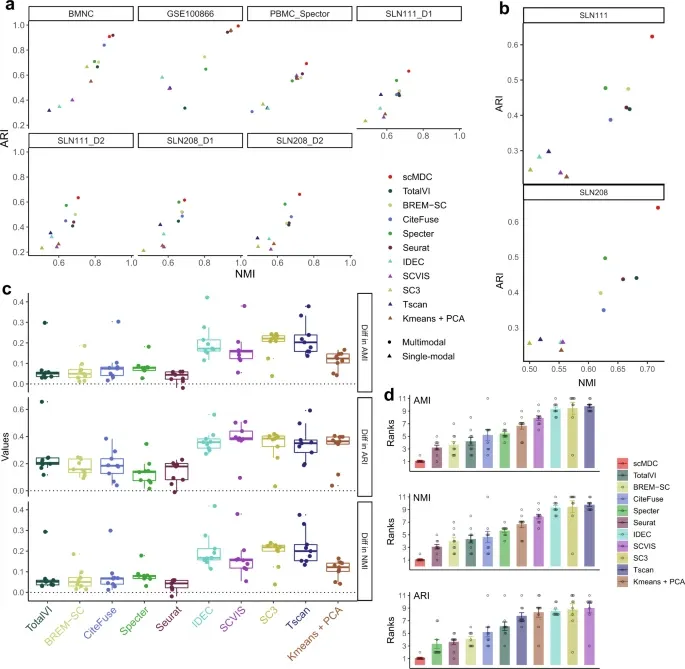
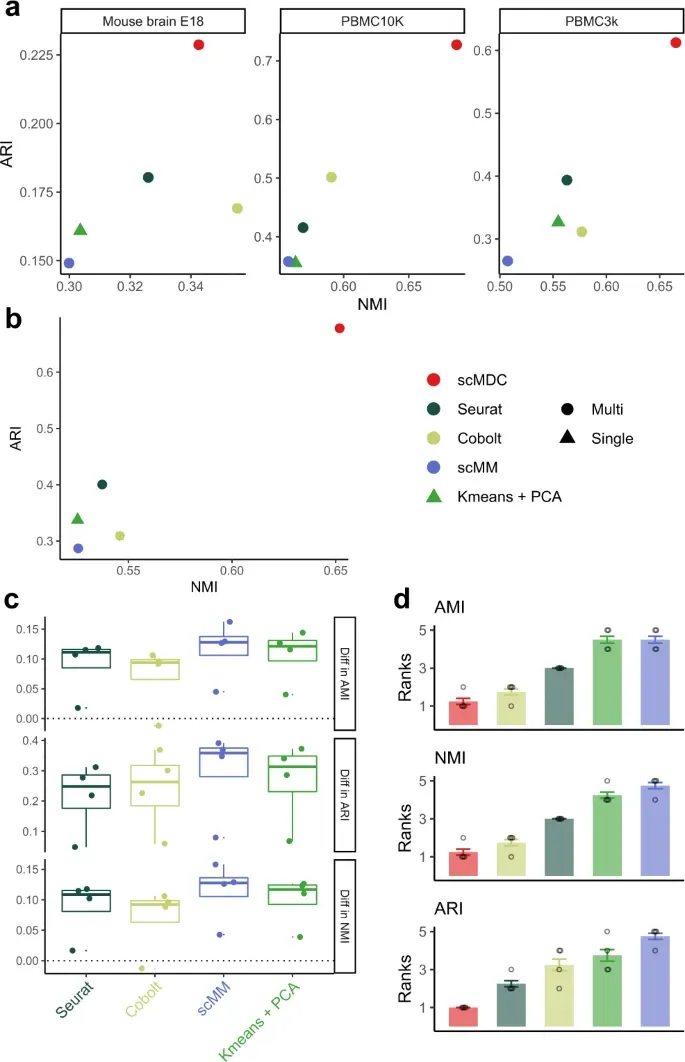
Results
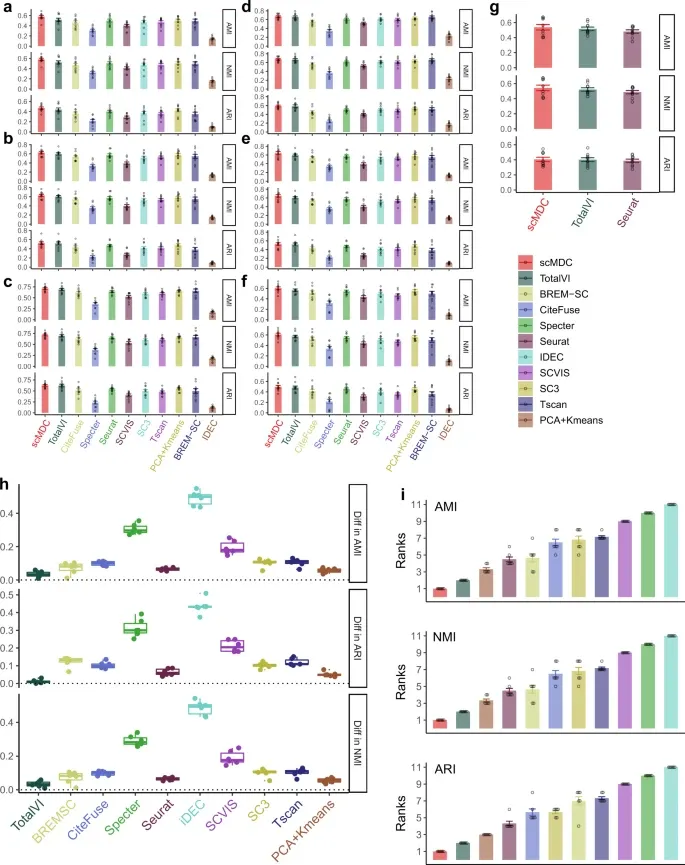
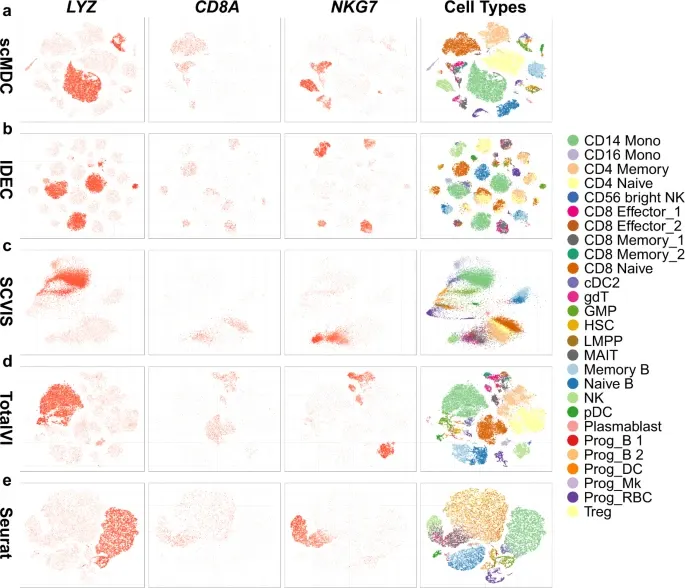
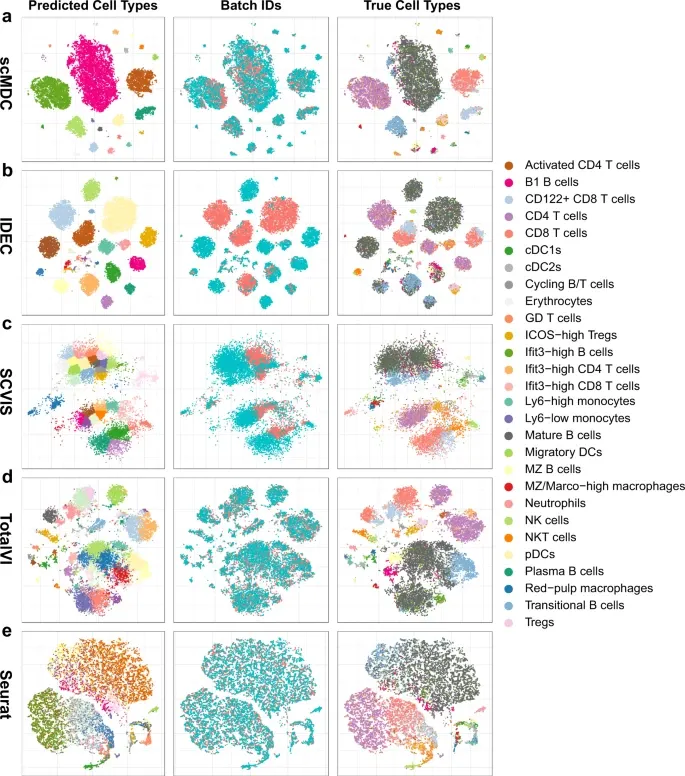
1. [CITE-seq dataset] 다른 clustering method와 비교
Evaluation metric: ARI, AMI, NMI