•
Access to data: GSE149383
Novelty
•
Drug dose를 고려한 time-series single-cell RNA-seq 데이터 생산
•
LINCS 데이터를 활용한 drug combination identification pipeline 제안
◦
Statistical test 기반
•
Hypothesis : Drug tolerance는 co-existence of heterogenous subpopulation 때문이다
•
Research question :다양한 subpopulation을 동시에 억제할 수 있는 drug combination을 찾는 다면 효과적인 치료가 되지 않을까?
•
keywords
: cancer cell subpopulation, drug tolerant vs sensitive cells, Markers for drug tolerant cell populations
Background
•
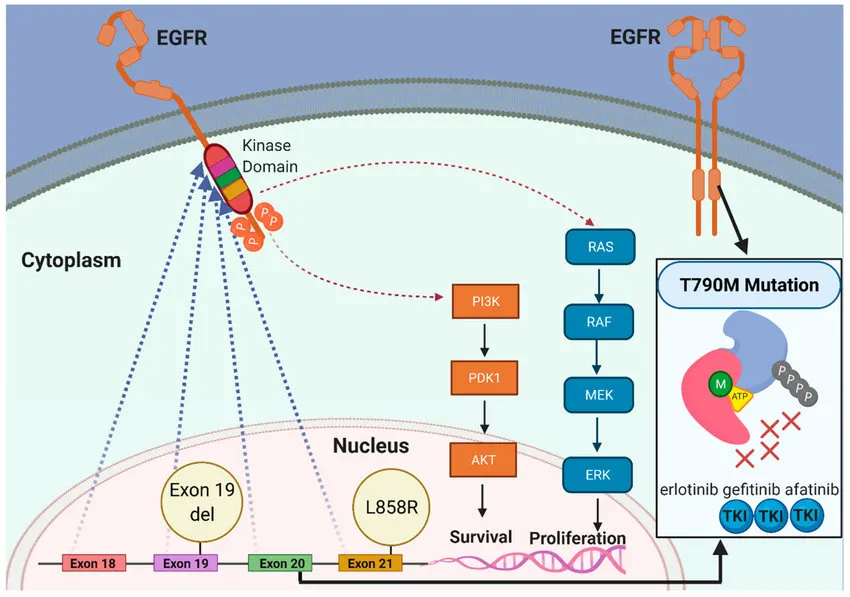
EGFR mutation-positive Non-small-cell lung carcinoma (NSCLC)
◦
NSCLC cell line: PC9 cell line (Exon 19 deletion in EGFR gene)
◦
Targets
▪
Epidermal growth factor receptor (EGFR)
T790M “gatekeeper” mutation in EGFR, which reportedly pre-exists or develops after several months of continuous treatment
◦
Drugs (표적 치료제)
Erlotinib
: targets and inhibits EGFR
Osimertinib
: targets and inhibits EGFR
•
Significant variability in drug response at the level of individual cells
◦
Drug-resistant colonies derived from a single cell were deficient in many known erlotinib-resistance mechanisms and was quite surprising and suggested that different surviving cells employ distinct mechanisms.
◦
As genotyping and drug- sensitivity testing required high cell numbers, which usually could only be achieved after 12–16 weeks of expanding rare tolerant cells in culture, there was gap in analysis of genetic and epigenetic heterogeneity during the initial robust response to targeted therapy. → Exclusion of information regarding the earlier sequence of events
Goal
Inspection of cell-level drug response on earlier stage (~Day 19) after treatment
1.
Distinguishing drug-tolerant states
2.
Discovering unique cell subpopulations, likely reflecting functional heterogeneity of drug-tolerant cells
3.
Suggest drug combination with LINCS analysis (proposed by the authors)